
Here, we introduced SeqKit toolkit to address the need for efficient and facile manipulations of FASTA/Q files. With the increasing number of sequences being produced, processing efficiency has become critical. Moreover, some tools require dependencies or running environments for installation or are only available for specific operating systems, which render them less user friendly. However, most of these programs implement only some of the above functions necessary for common manipulation and are not efficient for large files. Many tools are available for the manipulation of FASTA/Q files, including fasta_utilities, fastx_toolkit, pyfaidx, seqmagick and seqtk. Most of these scripts are not well organized or documented and are not reusable by other researchers. However, researchers, especially beginners, repeatedly write scripts for common purposes such as extracting sequences by using an identifiers (IDs) list file. The simplicity of the FASTA/Q formats makes them easy to be parsed and manipulated with programming languages like Python and Perl. Common manipulations of FASTA/Q files include converting, cleaning, searching, filtering, deduplication, splitting, shuffling, and sampling. FASTA was introduced first in FASTA software, and FASTQ was originally developed at the Wellcome Trust Sanger Institute.
įASTA and FASTQ are basic and ubiquitous text-based formats for storing nucleotide and protein sequences. SeqKit is open source and available on Github at.

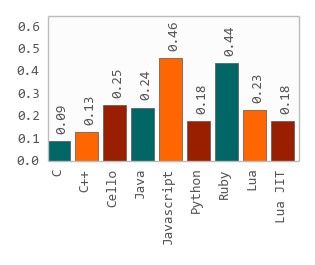
The efficiency and usability of SeqKit enable researchers to rapidly accomplish common FASTA/Q file manipulations. SeqKit demonstrates competitive performance in execution time and memory usage compared to similar tools.
#Klib library mac#
SeqKit provides executable binary files for all major operating systems, including Windows, Linux, and Mac OSX, and can be directly used without any dependencies or pre-configurations. This paper describes a cross-platform ultrafast comprehensive toolkit for FASTA/Q processing. Furthermore, the complicated installation process of required packages and running environments can render these programs less user friendly. Existing tools only implement some of these manipulations, and not particularly efficiently, and some are only available for certain operating systems. Common manipulations of FASTA/Q file include converting, searching, filtering, deduplication, splitting, shuffling, and sampling. FASTA and FASTQ are basic and ubiquitous formats for storing nucleotide and protein sequences.


 0 kommentar(er)
0 kommentar(er)
